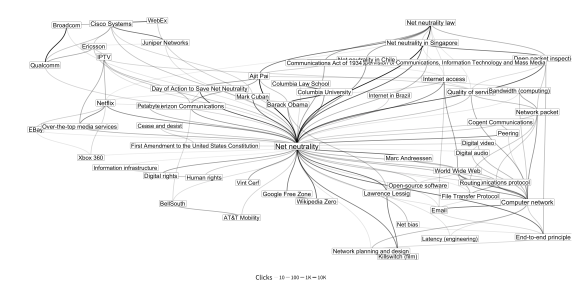
Diagram by Mikhail Popov/Wikimedia Foundation, CC BY-SA 4.0.
To go together with the announcement post for the monthly Wikimedia Clickstream releases, we wanted to create a graphic that would showcase the network aspect of the dataset:
In this post, you’ll learn a little bit about graph theory, how to import the clickstream data into the programming language and statistical software “R”, how to convert it into a graph, and how to visualize it to create the figure above. Some familiarity with R is required to follow along, but a list of free resources is available at the bottom for those who are interested in learning it.
Setting up
First, you’ll need R installed on your system. It is available on Comprehensive R Archive Network (CRAN) for Linux, macOS, and Windows. Then you will need to install some additional packages which are available on CRAN:
install.packages(c("igraph", "ggraph"))
If trying to attach the ggraph package with library(ggraph) gives you an error, you need to install the development version of ggplot2:
# install.packages("devtools")
devtools ::install_github("tidyverse/ggplot2")
Quick vocabulary lesson
A graph is a structure containing a set of objects where pairs of objects called vertices (also called nodes) are connected to each other via edges (also called links). Those edges represent relationships and they may be directed or undirected (mutual), and they may have weights. Two vertices that are endpoints of the same edge are adjacent to each other, and the set of vertices adjacent to the vertex V is called a neighborhood of V.
For example, if we have a group of people and we know how much each person trusts someone else, we can represent this network as a directed graph where each vertex is a person and each weighted, directed edge between vertices A and B is how much person A trusts person B (if at all!). Each person who A trusts or who trusts A collectively form a neighborhood of people adjacent to A.
There are additional terms to describe different properties of graphs (such as when a path of edges exists between any two vertices), but they are outside the scope of this post. If you are interested in learning more, please refer to the articles on graph theory and graph theory terms.
Reading data
First, we have to grab the dataset from Wikimedia Downloads and uncompress it before reading it into R. Once it’s ready, we can use the built-in utility for importing it. In this case we are working with the English Wikipedia clickstream from November 2017:
enwiki <- read.delim(
"clickstream-enwiki-2017-11.tsv",
sep = "\t",
col.names = c("source", "target", "type", "occurrences")
)
Due to the size of the dataset, this will take a while. To speed up this step (especially if something goes wrong and you have to restart your session), we recommend using readr package. Unfortunately we were unable to read in the original file with readr so we had to use the built-in utility to read the data first, write it out using readr::write_tsv (which performed all the necessary character escaping), and then read it with readr in subsequent sessions. If you have gzip, you can compress the newly created version and read in the compressed version directly:
enwiki <- readr::read_tsv("clickstream-enwiki-2017-11-v2.tsv.gz")
Working with graphs
Now that we have the data inside R, we have to convert it to a special object that we can then do all sorts of cool, graph-y things with. We accomplish this with the igraph network analysis library (which has R, Python, and C/C++ interfaces).
Although the data includes counts of users coming in from search engines and other Wikimedia projects, we restrict the graph to just clicks between articles and specify that the connections described by our data have a direction:
library(igraph)
g <- graph_from_data_frame(subset(enwiki, type == "link"), directed = TRUE)
We use the make_ego_graph function to construct a sub-graph that is a neighborhood of articles adjacent to our article of choice. In igraph, we can use the functions V and E to get/set attributes of vertices and edges, respectively, so we also create additional attributes for the vertices (such as the number of neighbors via degree)
sg <- make_ego_graph(g, order = 1, mode = "out", V(g)["Net_neutrality"])[[1]]
# Number of neighbors:
V(sg)$edges <- degree(sg)
# Labels where the spaces aren't underscores:
V(sg)$label <- gsub("_", " ", V(sg)$name, fixed = TRUE)
Because of the high number of articles in this neighborhood, to make a cleaner diagram we want to omit articles that have fewer than two neighbors — that is, a sub-sub-graph that only has vertices which have at least two edges:
ssg <- induced_subgraph(sg, V(sg)[edges > 1])
Visualization
In the data visualization package ggplot2 (and the “grammar of graphics” framework), you make a composition from layers that have geoms (geometric objects) whose aesthethics are mapped to data, potentially via scales such as size, shape, and alpha levels (also known as opacity/transparency), and color. The extension ggraph works on graph objects made with igraph and allows us to use a familiar language and framework for working with network data.
In order to draw a graph, the vertices and edges have to be arranged into a layout via an algorithm. After trying various algorithms, we settled on the Davidson-Harel layout to visualize our sub-sub-graph:
set.seed(42) # for reproducibility
ggraph(ssg, layout = "dh") +
geom_edge_diagonal(aes(alpha = log10(occurrences))) +
scale_edge_alpha_continuous("Clicks", labels = function(x) { return(ceiling(10 ^ x)) }) +
geom_node_label(aes(label = label, size = edges)) +
scale_size_continuous(guide = FALSE) +
theme_graph() +
theme(legend.position = "bottom")
We map the opacity of the edge geoms to the count of clicks to show volume of traffic between the articles (using a log10 transformation to adjust for positive skew). We also map the size of the label geoms to the number of neighbors, so the names of articles with many adjacent articles show up bigger.
Note that due to the way many graph layout algorithms work — where a random initial layout is generated and then iterated on to optimize some function — if you want reproducible results you need to specify a seed for the (pseudo)random number generator.
If you would like to learn how to work with ggplot2‘s geoms, aesthethic mappings, and scales, UCLA’s Institute for Digital Research and Education has a thorough introduction. I also recommend the data visualization chapter from R for Data Science.
Parting words
I hope this was helpful in getting you started with Wikimedia Clickstream data in R, and I look forward to seeing what the community creates with these monthly releases! Please let us know if you are interested in technical walkthrough posts like this.
Learning R
If you are interested in learning R, here are some free online resources:
Beginner’s guide to R by Sharon Machlis
DataCamp’s Introduction to R
Code School’s Try R
edX’s Introduction to R for Data Science
swirl: Learn R, in R
R for Data Science by Garrett Grolemund and Hadley Wickham
RStudio webinars
Mikhail Popov, Data Analyst, Reading Product
Wikimedia Foundation

Can you help us translate this article?
In order for this article to reach as many people as possible we would like your help. Can you translate this article to get the message out?
Start translation