Since October 2012, Wikidata has evolved a lot to become one of the most important open knowledge graphs, providing semantic knowledge about various topics in multiple languages. This effort includes the development of quality information for Biomedicine that can be reused for clinical decision support among other very important tasks.
In 2019, we conducted a research study to assess the coverage of health-related information in Wikidata and we found that it lacks support of various important types of information and that a significant set of biomedical relations has a limited precision and is not linked to references. Despite the use of crowdsourcing and human editing, the situation does not evolve as it should be. We needed a hack to change all the game.
MeSH Keywords as a valuable resource
MeSH (Medical Subject Headings) keywords play a pivotal role in the realm of biomedical knowledge representation, making them a valuable resource in various aspects of healthcare research and practice. It is composed of a heading providing the main topic of a research paper and a qualifier identifying the facet of the topic that is discussed by the paper.

Created and maintained by the National Library of Medicine (NLM), MeSH keywords provide a standardized vocabulary for indexing, cataloging, and searching for biomedical and health-related information. Here are some key reasons why MeSH keywords are considered a valuable resource:
Standardized Terminology
MeSH keywords offer a standardized and structured way to describe biomedical concepts. Each keyword is associated with a unique identifier, facilitating interoperability and data integration across various biomedical databases and systems. This standardization ensures that researchers, healthcare professionals, and data scientists are speaking the same language when referring to specific medical topics, which is crucial in a field where precise terminology is paramount.
Improved Search and Retrieval
MeSH keywords significantly enhance information retrieval in biomedical databases such as PubMed. Researchers and healthcare practitioners can use MeSH terms to refine their searches, ensuring that they find highly relevant articles and resources. This precision in search and retrieval expedites literature reviews, clinical decision-making, and evidence-based practice.
Hierarchy and Relationships
MeSH keywords are organized into a hierarchical structure. This hierarchy enables researchers to navigate from broader concepts to more specific ones, making it easier to explore related topics and delve deeper into a subject area. This feature is especially beneficial for researchers seeking to understand complex medical phenomena or clinicians aiming to grasp the broader context of a particular condition.
Facilitation of Data Annotation
In the context of open knowledge graphs like Wikidata, MeSH keywords are instrumental in annotating and linking biomedical concepts. By associating MeSH terms with specific entities or topics, it becomes possible to create structured and interconnected knowledge representations. These annotations not only serve as a basis for data integration but also enable advanced semantic querying, classification, and reasoning.
Enabling Multidisciplinary Collaboration
MeSH keywords bring together professionals from diverse backgrounds, including medicine, biology, pharmacology, and computer science. This shared terminology ensures that collaborative projects can effectively bridge the gap between clinical knowledge and technical expertise. Multidisciplinary teams can collaborate seamlessly to enrich, validate, and apply biomedical knowledge in innovative ways.
Research and Clinical Decision Support
The value of MeSH keywords extends beyond research; they are instrumental in clinical decision support systems. Healthcare providers can use structured MeSH terminology to access the latest medical literature and clinical guidelines, aiding them in diagnosing patients, determining treatment plans, and staying current with advancements in healthcare. MeSH keywords empower healthcare professionals to make informed decisions based on a robust foundation of medical knowledge.
Using MeSH Keywords for adjusting Wikidata
The integration of MeSH (Medical Subject Headings) keywords into Wikidata represents a significant leap forward in the effort to enhance and adjust the open knowledge graph for clinical and biomedical applications. MeSH keywords, meticulously curated and structured for biomedical content, provide a powerful framework for improving the quality, coverage, and relevance of Wikidata in the context of healthcare and clinical practice. The Project entitled “Adapting Wikidata to support clinical practice using Data Science, Semantic Web and Machine Learning” has been funded by the Wikimedia Foundation Research Fund to assess this direction. It was launched in August 2022 by the Data Engineering and Semantics Research Unit at the University of Sfax, Tunisia alongside the School of Data Science at the University of Virginia, United States of America and the Institute for Technological Innovation at the University of Pretoria, South Africa.
Identifying concerns about biomedical relations in Wikidata
In order to identify potentially inconsistent relations between Wikidata items aligned with MeSH taxonomy, we employ the Pointwise Mutual Information (PMI) metric for each relation. PMI serves as a corpus-derived measure of semantic relatedness, highlighting relations as significant when they surpass a predefined threshold, typically set at 2.
To calculate PMI using MeSH keyword associations, several values are required:
- N(x): The frequency of occurrences of the subject in PubMed.
- N(y): The frequency of occurrences of the object in PubMed.
- N(x,y): The count of associations between the subject and the object in PubMed.
- P: The total number of PubMed records available.
These essential values are used in the PMI calculation to assess the strength of relations between MeSH-aligned Wikidata items. This method can be scaled to any kind of statements, including non-relational ones, and adapted to be driven by search engines.
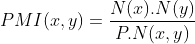
The PMI calculation for the 109,302 Wikidata relations between MeSH-aligned items reveals the following:
- 12,898 relations (11.8%) cannot be verified due to inaccurate MeSH ID data in Wikidata. You can find the list of these erroneous MeSH ID values at MeSH Verification Spreadsheet.
- 40,725 relations (37.2%) are likely to be incorrect and require verification by medical specialists as they fall below the predefined PMI threshold. The list of these semantic relations needing attention can be accessed at Relations Requiring Verification.
These resources provide detailed information for further analysis and validation of the identified issues in the MeSH-aligned Wikidata relations. Please note that several accurate non-biomedical relations can have PMI values less than 2 as PubMed does not efficiently cover scholarly information not related to medical practice.
Identifying new biomedical relations from PubMed
We identified the most common 5,000 MeSH keywords in PubMed and we studied the associations with them using PMI. This stage involve the computation of 25 million PMI values and consequently requires parallel computing and high computational capacities. Due to this computational complexity, we developed a data center at the University of Sfax thanks to the efforts of two contractors and the funding of the Wikimedia Foundation, the WikiCred Grant Initiative, and the Tunisian Ministry of Higher Education and Scientific Research among other institutions.
The Project’s Final Office Hour in French (September 30, 2023) featuring a demonstration of the project’s work and the data center. Please enable auto-translated closed captions in English for more context.
Thanks to these efforts, we successfully identified 835,111 new relations between the 5000 most common MeSH-aligned Wikidata items. These new relations are supported by PubMed references identified thanks to the PubMed Best Match sorting method. This number seems to be huge when compared to the current number of relations between MeSH-aligned Wikidata items. That is why we need a significant community of Wikidata contributors having robust medical knowledge to go through these relations and verify whether they are relevant or not. As well, we need to identify the Wikidata properties corresponding to these associations so that they can be added to Wikidata using QuickStatements or another batch upload tool. You can find the list of the significant associations between the 5000 most important MeSH keywords here.
Classifying new biomedical relations
In the classification process, we employed qualifiers associated with both the subject and object of each relation across 20 publications or fewer. This data served to create an association matrix that links subject qualifiers to object qualifiers for each statement. Following this, classification is conducted using a dense neural network, responsible for assigning both the relevant Wikidata Property for the relation and determining the appropriate type of Wikidata Property for assignment to the relation.
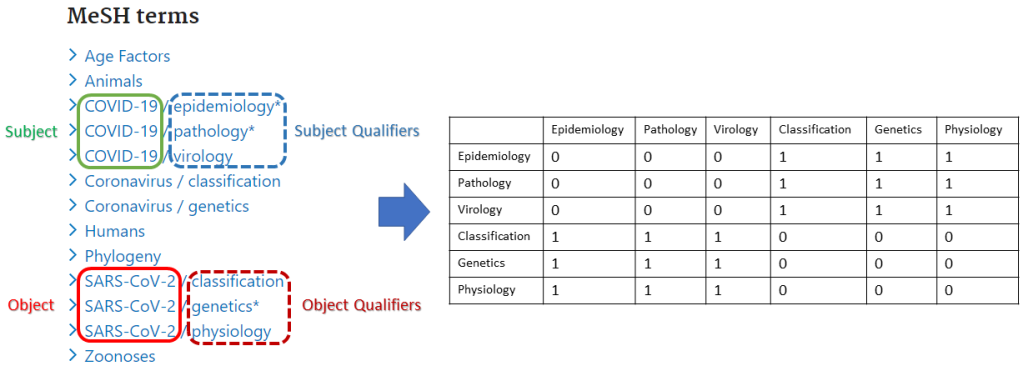
In cases where a conflict arises between the returned Wikidata property and the assigned property type, the classification is deemed incorrect, and its result is not ascribed to the respective relation. The classification process has been proven to be accurate at a rate of 89.40% for superclass-based classification and 75.32% for relation type-based classification. The joint verification of the superclass-based classification and the relation type-based classification has been identified as efficient in removing 93.1% of the mistaken classification outputs. The source code for the supervised classification algorithm is available at MeSH2Wikidata. While this method demonstrates efficiency, there is no guarantee that it can classify a significant portion of new relations, primarily because several MeSH associations lack qualifiers. That is why the contribution of the Wikidata community is required to classify these relations.
Conclusion
We are confident that our efforts mark one of the initial strides toward revolutionizing the application of Artificial Intelligence in Wikimedia Projects. We look forward to collaborating with Wikimedia Deutschland and the Wikimedia Foundation Research and Development teams to advance this undertaking. Their collaboration with our consortium is crucial to expand the scope of our accomplishments and establish a sustainable solution that can elevate Wikidata into a dependable and versatile multidisciplinary knowledge graph.

Can you help us translate this article?
In order for this article to reach as many people as possible we would like your help. Can you translate this article to get the message out?
Start translation